
Damien Wilburn
Assistant Professor
880 Biological Sciences Building
484 W. 12th Avenue
Columbus, OH 43210
Areas of Expertise
- Biochemistry
Biography
Damien Wilburn attended the University of Louisville where he received his B.Sc. in Biology and Mathematics in 2009 as well as his Ph.D. in Biochemistry and Molecular Biology in 2014. Beginning in 2007 as an undergraduate research scholar and continuing into his PhD as an NSF Graduate Research Fellow, he studied the structural dynamics, translational regulation, and molecular evolution of hypervariable courtship pheromones in lungless salamanders under the mentorship of Drs. Richard and Pamela Feldhoff. This work fostered a broader interest in sexual selection and reproductive biology which motivated his interdisciplinary postdoctoral research in sperm-egg interactions at the University of Washington (UW) under Dr. Willie Swanson, with co-mentorship from Drs. Rachel Klevit (UW; NMR) and Brian Searle (OSU; MS and deep learning). During his postdoctoral tenure, he was awarded a NIGMS F32 fellowship and NICHD K99/R00 career award, in addition to being named the 2017 Outstanding Postdoc Mentor by the UW Graduate School. He joined the OSU Department of Chemistry and Biochemistry in August 2022.
Research Overview
In animals, reproduction-associated genes have evolved at extraordinary rates (often faster than immune genes) which has produced tremendous diversity in molecular structure and function. Co-evolution of sperm and egg proteins contributes to species-specific gamete recognition, but the biophysical mechanisms of how gametes fuse with such exquisite specificity remain poorly defined. My laboratory studies the biochemistry of animal fertilization in three models: humans, lungless salamanders, and marine abalone. Each system offers unique experimental opportunities for understanding core mechanisms of animal fertilization, the evolution of reproductive barriers, and possible drivers of human infertility. For known reproductive proteins, biophysical properties and molecular interactions are studied using a range of biochemical tools including multidimensional NMR that can measure structural dynamics and weak binding interactions common to rapidly evolving proteins. But rapid sequence evolution challenges homology-based gene identification/annotation such that not all reproductive proteins have been identified, even in well-studied species like humans, and my laboratory uses mass spectrometry-based quantitative proteomics to identify candidate fertilization proteins. New software tools and statistical models are developed as needed for analyzing biophysical, mass spectrometry, and evolutionary genomic data.
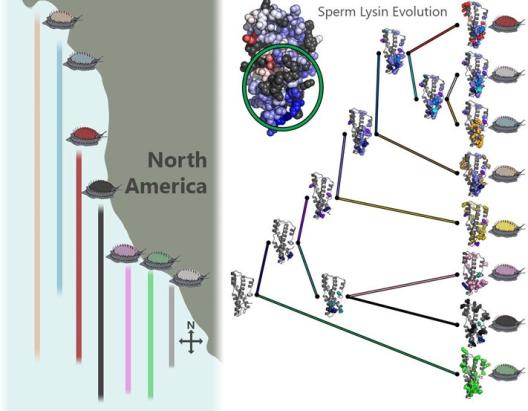
Abalone
The marine mollusk abalone is the most well characterized animal model of fertilization. As a broadcast spawning marine invertebrate with external fertilization, both sperm and egg can be easily collected in enormous quantities for biochemical and functional characterization (e.g., more than 1 gram of the major sperm protein lysin can be purified from a single male abalone). Off the North American Pacific coast exist 7 abalone species (whose common names are often based on the color of their shells) with overlapping habitat ranges that creates substantial opportunity for hybridization; Despite hybrids being viable in the laboratory they are rarely observed in the wild, with molecular recognition between egg and sperm being one of the major factors that restricts heterospecific mating. Compared to mammalian counterparts, the abalone sperm proteome is remarkably simple and consists largely of only three proteins: lysin, sp18, and FITZAP. Lysin interacts with ZP-N domains of egg VERL to permeate the vitelline envelope (homologous to the mammalian zona pellucida, ZP). Sp18 is an ancient paralog of lysin with a similar tertiary structure despite low sequence similarity that is highly fusogenic and likely facilitates sperm-egg fusion. FITZAP is a small intrinsically disordered protein that enables packaging of lysin and sp18 inside the sperm acrosome at concentrations nearing one molar. As all three abalone sperm proteins are small and highly soluble, they are well suited for structural analysis by solution NMR. One barrier to abalone hybridization is the rapid co-evolution of sperm lysin and egg VERL which has resulted in weak but species-specific interactions between the gamete recognition molecules. On-going work in the lab is focused on characterizing the interactions between egg ZP-N domains, structural mechanisms of lysin-mediated egg coat dissolution, and the structure-function dynamics of sp18 with the oocyte plasma membrane.
Humans
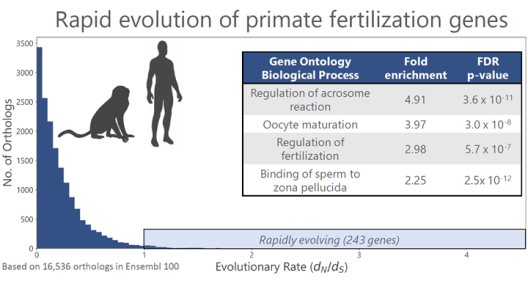
The biochemical underpinnings of human fertilization are largely a molecular mystery. In contrast to marine invertebrates such as abalone, the proteomic composition of mammalian sperm is substantially more complex in composition and functional redundancy, and there are only two known pairs of interacting sperm-egg proteins in humans. Rapid evolution and lineage-specific diversification of gametic proteins also limits the utility of genetic studies in rodent models for understanding human fertilization. My laboratory integrates quantitative mass spectrometry of human gametes with molecular evolutionary analyses and deep learning to identify new interacting pairs of fertilization proteins that are characterized using structural and biophysical techniques.
Salamanders
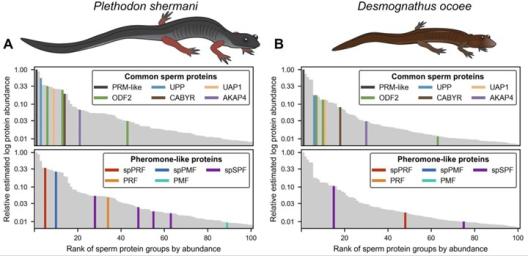
Lungless salamanders (family Plethodontidae) are classic models of reproductive behavior and mating. Male and female salamanders perform stereotyped courtship behaviors that culminate in external sperm transfer for internal fertilization, and such behaviors include the “tail straddling walk” that has persisted for ~130 million years of salamander evolution. In the majority of plethodontid species, male salamanders deliver non-volatile protein courtship pheromones that modulate female mating behavior. These pheromones have been co-opted from numerous gene families and experienced rapid evolution, presumably in response to coevolutionary pressures from female receptors. Three pheromone types have been chemically purified and experimentally demonstrated to alter female mating behavior: PRF, PMF, and SPF. Remarkably, recent analysis by quantitative mass spectrometry identified paralogs of all three pheromone families as major sperm proteins that are hypothesized to play essential roles in salamander fertilization. As the most speciose family of salamanders (496 of 766 species), plethodontid salamanders have experienced expansive radiations throughout the North and Central American coasts where many species are divided along ecological gradients (e.g. elevation) with varying levels of hybridization and genetic introgression. Such hybrid zones provide natural experiments to explore how new reproductive barriers – and ultimately speciation – may evolve within homologous proteins found at both pre-mating (pheromone) and post-mating (sperm) molecular interactions.
Select Publications
The full and up-to-date list of publications can be found in Google Scholar.
Wilburn, D.B., Kunkel, C.L, Feldhoff, R.C., Feldhoff, P.W., and Searle, B.C. (2022). Recurrent co-option and recombination of cytokine and three proteins in multiple reproductive tissues throughout salamander evolution. Frontiers in Cellular and Developmental Biology, 10: 828947. doi: 10.3389/fcell.2022.828947
Wilburn, D.B., Tuttle, L.M., Klevit, R.E., and Swanson, W.J. (2019). Indirect sexual selection drives rapid sperm protein evolution in abalone. eLife, 8: e52628. doi:10.7554/eLife.52628
Wilburn, D.B. and Feldhoff, R.C. (2019). An annual cycle of gene regulation in the red-legged salamander mental gland: from hypertrophy to expression of rapidly evolving pheromones. BMC Developmental Biology, 19: 10. doi:10.1186/s12861-019-0190-z
Wilburn, D.B., Tuttle, L.M., Klevit, R.E., and Swanson, W.J. (2018). Solution structure of sperm lysin yields novel insights into molecular dynamics of rapid protein evolution. Proceedings of the National Academy of Sciences, 115(6): 1310-1315. doi:10.1073/pnas.1709061115
Wilburn, D.B., Doty, K.A., Chouinard, A.J., Eddy, S.L., Woodley, S.K., Houck, L.D., and Feldhoff, R.C. (2017). Olfactory effects of a hypervariable multicomponent pheromone in the red-legged salamander, Plethodon shermani. PLOS ONE 12(3): e0174370. doi:10.1371/journal.pone.0174370
Wilburn, D.B., Eddy, S.L., Chouinard, A.J., Arnold, S.J., Feldhoff, R.C., and Houck, L.D. (2015). Pheromone isoform composition differentially affects female behaviour in the red-legged salamander, Plethodon shermani. Animal Behaviour. 100: 1-7. doi:10.1016/j.anbehav.2014.10.019
Wilburn, D.B., Bowen, K.E., Doty, K.A., Arumugam, S., Lane, A.N., Feldhoff, P.W., and Feldhoff, R.C. (2014). Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable vertebrate pheromone from the red-legged salamander. PLOS ONE 9(5): e96975. doi:10.1371/journal.pone.0096975
Wilburn, D.B., Bowen, K.E., Gregg, R.G., Cai, J., Feldhoff, P.W., Houck, L.D., and Feldhoff, R.C. (2012). Proteomic and UTR analysis of a rapidly evolving hypervariable family of vertebrate pheromones. Evolution, 66(7): 2227-2239. doi: 10.1111/j.1558-5646.2011.01572.x
