
Amanda Hummon
Professor
414 Biomedical Research Tower
460 W 12th Ave
Columbus, OH 43210
Areas of Expertise
- Analytical
- Biochemistry
Biography
Amanda Hummon earned her A.B. in chemistry at Cornell University in 1999 with honors. She completed her graduate studies in analytical chemistry at the University of Illinois, Urbana-Champaign, in the laboratory of Prof. Jonathan V. Sweedler. Her thesis work focused on the development of mass spectrometric and bioinformatic strategies to predict and identify neuropeptides. She received her Ph.D. in 2004. Amanda participated in the annotation of the newly sequenced honeybee genome as a post-doctoral fellow in the laboratories of Prof. Gene E. Robinson and Prof. Sandra L. Rodriguez-Zas at the University of Illinois from 2004-2005. The focus of her research was constructing a methodology to utilize detected gene products to decipher an unannotated genome. From 2005-2009, Amanda was a Sallie Rosen Kaplen Post-Doctoral Fellow at the National Cancer Institute in the laboratory of Dr. Thomas Ried. During her time in the Ried lab, she utilized RNA interference screening techniques followed by microarray analysis to elucidate genes that regulate the viability of colorectal cancer cells.
In 2009, she began her independent career in the Department of Chemistry and Biochemistry at the University of Notre Dame and moved her research program to the Department of Chemistry and Biochemistry at The Ohio State University in January 2018. She has been recognized with an NSF CAREER award (2014), a Society for Analytical Chemists of Pittsburgh Starter Grant Award (2011), a Rising Star Award from the American Chemical Society (2016) and the Presidential Early Career Award for Scientists and Engineers (PECASE) in 2019. In 2020, she was awarded a Fulbright Scholar Award and spent a semester as a Visiting Professor at the Maastricht MultiModal Molecular Imaging Institute at Maastricht University in the Netherlands.
Research Overview:
Our research interests lie at the intersection of analytical chemistry and chemical biology, with a focus on cancer biology. Cancer is a complex disease, requiring sophisticated and systematic strategies to deliver knowledge that will lead to improved treatment options for patients. Individuals trained in a wide range of scientific topics and possessing a substantial breadth of knowledge are best positioned to tackle this challenge. In the Hummon Research Group, we develop analytical methods to evaluate both the transcriptome and the proteome in cancer cells, while exploring the deregulation in cancer-associated signal transduction pathways.
Cancer arises from insults to the genome. With genomic damage, the expression levels of genes are altered from their normal state. Changes in the genome, transcriptome and proteome have been found to be highly conserved among samples from adenomas to carcinomas to metastases. Because genetic changes are commonly repeated among cancer patients, a better understanding of which genes, transcripts, and proteins are affected could have broad health implications. Therefore, the best way to understand the molecular underpinnings of cancer is to dissect the deregulated pathways that are contributing to the cancer phenotype, identify the aberrantly expressed genes and their products, and decipher their effect on downstream targets.
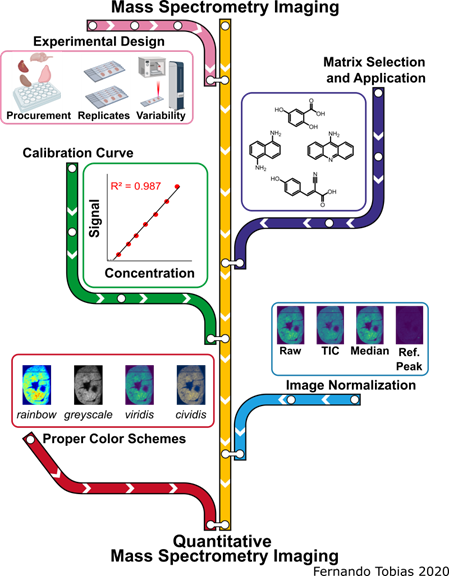
Biomolecule Visualization and Quantification Using Mass Spectrometry Imaging
The pink line represents experimental design and sources of variability, the purple line signifies the matrix selection process and application methods, the green line represents creating calibration curves for qMSI, the blue line symbolizes the image normalization aspect, while the red line represents the selection process for the proper color scheme of imaging data. The MS images depicted in this scheme are from the same murine heart section in a different color scheme and when the signal was normalized by different methods.
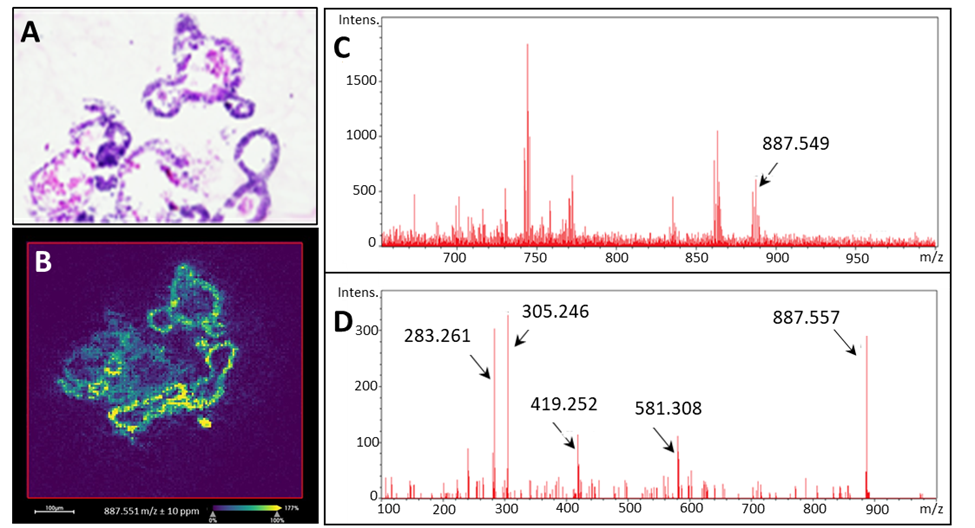
Organoid Analysis
Organoids are cell cultures that are derived from stem cells and can be grown from any organ in the body. We are developing approaches to map the endogenous and exogenous molecular species in these valuable samples by mass spectrometry imaging. For example, a lipid species (PI(18:0_20:3), m/z 887.55) is mapped and identified by MS/MS assessment.
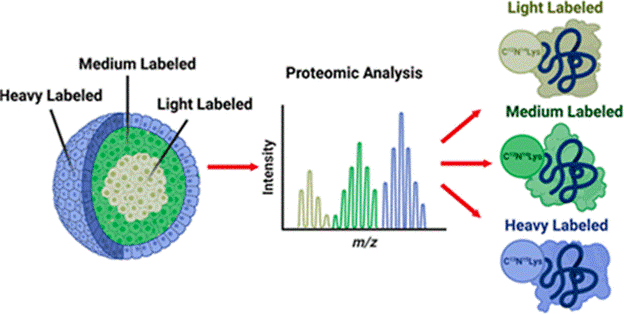
Proteomics: Spatial SILAC in Cellular Layers of the Spheroid
Stable isotopic labeling of amino acids in cell culture (SILAC) is a metabolic labeling method that identifies the origin of a protein in multiplexed experiments. By pulsing different heavy amino acids into cell culture media at set time points during spheroid growth, we can discretely label the cellular layers that form in these tumor models. Proteins from the necrotic core, quiescent middle layer, and proliferative outer layer of spheroids can be distinguished and quantified in a proteomic analysis, generating valuable information about chemotherapeutics’ effect on different cellular populations.
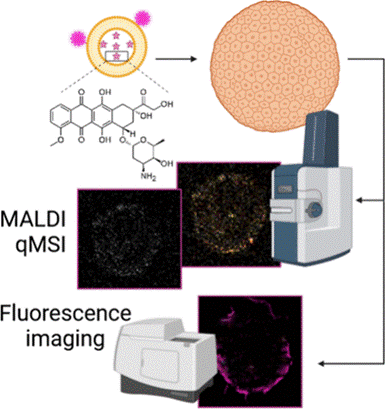
Imaging Mass Spectrometry Analysis of Liposomal Drug Delivery Systems
Cancer chemotherapeutics often fail to reach all diseased cells. To help solve this problem, researchers are investigating novel drug delivery systems. Liposomes are an attractive option due to their low toxicity, high biocompatibility, and potential to carry a large amount of a drug to the tumor site, all while avoiding being eliminated from the body. Using MALDI-IMS and fluorescence microscopy we analyze the spatial distribution of liposomal drugs in comparison to their free drug counterpart within spheroids. We also use click chemistry to modify the lipids and track their distributions by fluorescence.
Recent Publications
Sekera, E.R.; Rosas, L.; Holbrook, J.H.; Angeles, Q.D.; Khaliullin, T.; Rojas, M.; Mora, A.L.; Hummon, A.B. “Single Cell MALDI-MS Imaging of Lipids and Proteins in Senescent Fibroblasts” Journal of the American Society for Mass Spectrometry. 2024, 35(12) 2815-2823. PMID: 39476364
Fries, B. D.; Hummon, A.B. “FAS inhibited Proteomics and Phosphoproteomics Profiling of Colorectal Cancer Spheroids Shows Activation of Ferroptotic Death Mechanism” Journal of Proteome Research. 2024. 23(9) 3904-3916. PMID: 39079039
Lopez, A.; Holbrook, J.H.; Kemper, G.E.; Lukowski, J.K.; Andrews, W.T.; Hummon, A.B. “Tracking Drugs and Lipids: Quantitative Mass Spectrometry Imaging of Liposomal Doxorubicin Delivery and Bilayer Fate in Three-Dimensional Tumor Models” Analytical Chemistry. 2024. 96(22) 9254-9261. PMID: 38778440
Sekera, E.R.; Akkaya-Colak, K.B; Lopez, A.; Mihaylova, M.M.; Hummon, A.B. “Mass Spectrometry Imaging and Histology Workflow for the Analysis of Budding Intestinal Organoids” Analytical Chemistry. 2024. 96(10) 4251-4258. PMID: 38427328
Holbrook, J.H.; Kemper, G.; Hummon, A.B. “Quantitative Mass Spectrometry Imaging: Therapeutics & Biomolecules” Chem Comm. 2024 60(16) 2137-2151. PMID: 38284765
Shannon, A.E. Boos, C.E.; Searle, B. C.; Hummon, A. B. “Gas-phase Fractionation Data-Independent Acquisition analysis of 3D Cocultured Spheroid Tumor Model Reveals Altered Translational Processes and Signaling using Proteomics” Journal of Proteome Research, 2024. 23(8) 3188-3199.PMID: 38412258
Fries, B.D.; Tobias, F.; Wang, Y.; Holbrook, J.; Hummon, A.B. “Lipidomics Profiling Reveals Differential Alterations after FAS Inhibition in 3D Colon Cancer Cell Culture Models” Journal of the American Society for Mass Spectrometry. 2024, 23(8):2919-2933. PMID: 38063332
